Adding HiBiT Tag to an Endogenous Gene Using CRISPR
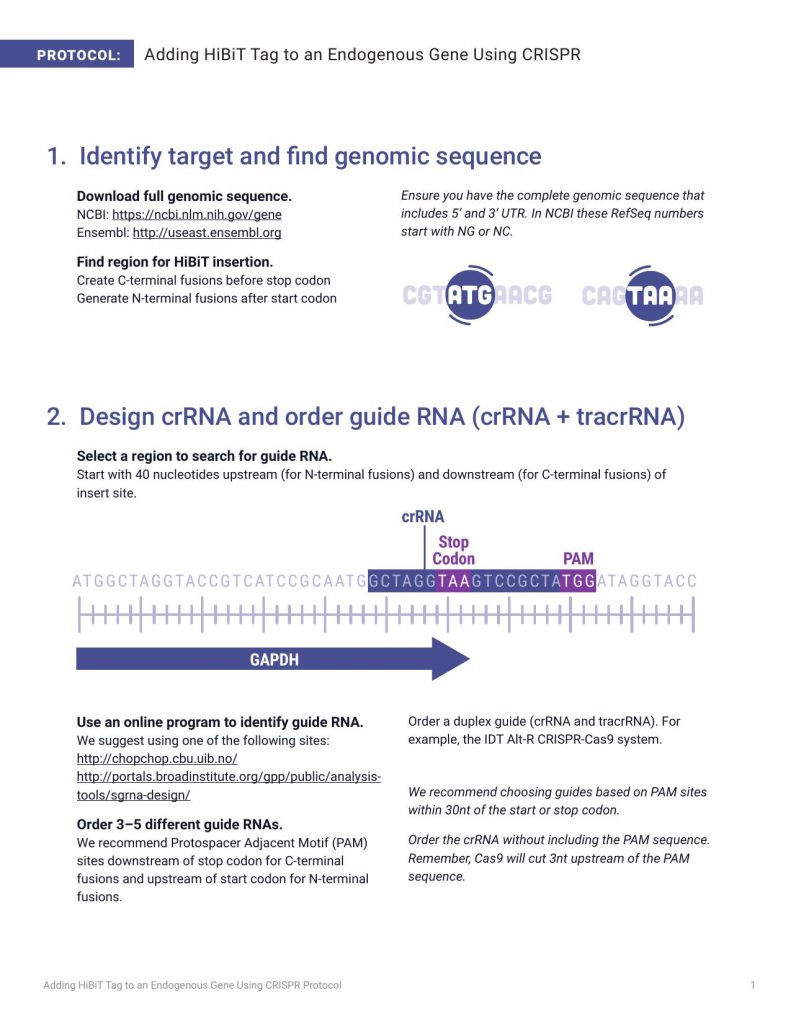
PROTOCOL: Adding HiBiT Tag to an Endogenous Gene Using CRISPR 2. Design crRNA and order guide RNA (crRNA + tracrRNA) Select a region to search for guide RNA. Start with 40 nucleotides upstream (for N-terminal fusions) and downstream (for C-terminal fusions) of insert site. Use an online program to identify guide RNA. We suggest using one of the following sites: http://chopchop.cbu.uib.no/ http://portals.broadinstitute.org/gpp/public/analysistools/sgrna-design/ Order 3–5 different guide RNAs. We recommend Protospacer Adjacent Motif (PAM) sites downstream of stop codon for C-terminal fusions and upstream of start codon for N-terminal fusions. Order a duplex guide (crRNA and tracrRNA). For example, the IDT Alt-R CRISPR-Cas9 system. We recommend choosing guides based on PAM sites within 30nt of the start or stop codon. Order the crRNA without including the PAM sequence. Remember, Cas9 will cut 3nt upstream of the PAM sequence. 1. Identify target and find genomic sequence Download full genomic sequence. NCBI: https://ncbi.nlm.nih.gov/gene Ensembl: http://useast.ensembl.org Find region for HiBiT insertion. Create C-terminal fusions before stop codon Generate N-terminal fusions after start codon Ensure you have the complete genomic sequence that includes 5’ and 3’ UTR. In NCBI these RefSeq numbers start with NG or NC. Adding HiBiT Tag to an Endogenous Gene Using CRISPR Protocol 1 14509MA 14509MA 14510MA Stop Codon PAM crRNA GAPDH ATGGCTAGGTACCGTCATCCGCAATGGCTAGGTAAGTCCGCTATGGATAGGTA CC 3. Design and order HiBiT Donor DNA template 4. Deliver guide RNA, donor DNA and Cas9 Find regions of identity for donor DNA. Use 50–80 nucleotides upstream and downstream of insert site. Position the HiBiT sequence symmetrically between homology arms. This facilitates the CRISPR homologuous recombination. Prepare ribonucleoprotein (RNP) complex. Anneal the crRNA to tracrRNA (IDT) to form guide RNA. Incubate purified Cas9 with guide RNA to form RNP complex. 1. Prepare 24µM tracrRNA:crRNA duplex. a. Combine the following in a PCR tube: b. Heat at 95℃ for 5 minutes and cool to room temperature on bench top. COMPONENT 100µM tracrRNA 100µM crRNA Nuclease-Free Duplex Buffer Total VOLUME 12µl 12µl 26µl 50µl COMPONENT 20µM Cas9 24µM trRNA:crRNA Total VOLUME 5µl 5µl 10µl 2. Prepare RNP complexes (sufficient for 200,000 cells). a. Combine 100pmol Cas9 and 120pmol tracrRNA:crRNA: Order donor DNA template. Request a single-stranded oligodeoxynucleotide (ssODN). For example, IDT Ultramer DNA Oligonucleotide. Obtain synthesis rights and access HiBiT sequence: www.promega.com/HiBiT-synthesis Introduce a silent mutation in the PAM sequence of the donor DNA template to avoid recutting. 2 Note: To avoid precipitation, add the Cas9 very slowly to the tracrRNA:crRNA and swirl with pipette tip. b. Allow RNP to form for 10–20 minutes at room temperature. Adding HiBiT Tag to an Endogenous Gene Using CRISPR Protocol 14511MA 5′ Exon Exon Exon 3′ UTR 3′ Single-Stranded Oligodeoxynucleotide ~80nt Homology Arm HiBiT Stop Codon Homology Arm 5′ 33nt HiBiT Sequence TGA ~80nt 3′ Deliver RNP and donor DNA to cell by electroporation. We have successfully electroporated commonly used cell lines (e.g., HeLa, U2OS, HepG2, HEK 293) suspended in Nucleofector Reagent using the cell line-specific program on the 4D-Nucleofector. We typically use 1µl of 100µM donor DNA template with 10µl of RNP for 20µl of cell suspension. We obtain highest efficiency using electroporation, however, lipid-based reagents designed for RNP transfection may be an alternative. Option 1: Use Lytic plate-based assay to confirm edited gene expression level. Take a sample of the edited cells 24–48 hours post-electroporation. Use Nano-Glo® HiBiT Lytic Detection System to assess luminescence for each guide RNA tested. Use unedited cells as negative control for background. Option 2: Use HiBiT Blotting to confirm expression of full-length protein. Take a sample of the edited cells and lyse with preferred buffer. Run samples on a gel, transfer proteins to membrane and detect using the Nano-Glo® HiBiT Blotting System. Use unedited cells as negative control for background. We recommend trypsinizing cells, resuspending to 200,000 cells/ml in growth medium and transferring 100µl (20,000 cells) per well of a 96-well plate. Transfer unused cells to a six-well plate to scale up for future experiments. Run lysates on gel Add Nano-Glo® HiBiT Blotting System reagents Transfer to membrane Detect luminescence 4. Deliver guide RNA, donor DNA and Cas9 (continued) 5. Validate editing event in cells (2 options) Adding HiBiT Tag to an Endogenous Gene Using CRISPR Protocol 3 Step 1 Step 1 Step 2 Step 3 Step 4 Step 2 Add Nano-Glo® HiBiT Detect luminescence Lytic detection reagent 14512MA Gl 14513MA oMax® Discover System 14514MA 14515MA 14516MA 14517MA PROMEGA CORPORATION • 2800 WOODS HOLLOW ROAD • MADISON, WI 53711-5399 USA • TELEPHONE 608-274-4330 www.promega.com • © 2017 PROMEGA CORPORATION • ALL RIGHTS RESERVED • PRICES AND SPECIFICATIONS SUBJECT TO CHANGE WITHOUT PRIOR NOTICE • PRINTED IN USA 09/17 • 38467583 • PART #GE777 DeWitt, M., Corn, J.E. and Carroll, D. (2017) Genome editing via delivery of Cas9 ribonucleoprotein. Methods Epub ahead of print. doi: http://dx.doi.org/10.1016/j.ymeth.2017.04.003 Detailed protocols from the Innovative Genomics Initiative at University of California–Berkeley at: https:// https://www.protocols.io/groups/innovative-genomics-institute Additional Resources
